Autonomous Tumor Tracking and Resection with Near Infrared Tumor Marking (FNIR) and the Smart Tissue Autonomous Robot (STAR)
PRINCIPAL INVESTIGATORS:
Alex Krieger, Department of Mechanical Engineering, University of Maryland
Martin Schnermann, Center for Cancer Research, National Cancer Institute
PH.D. STUDENT:
Jiawei Ge, Mechanical Engineering, University of Maryland
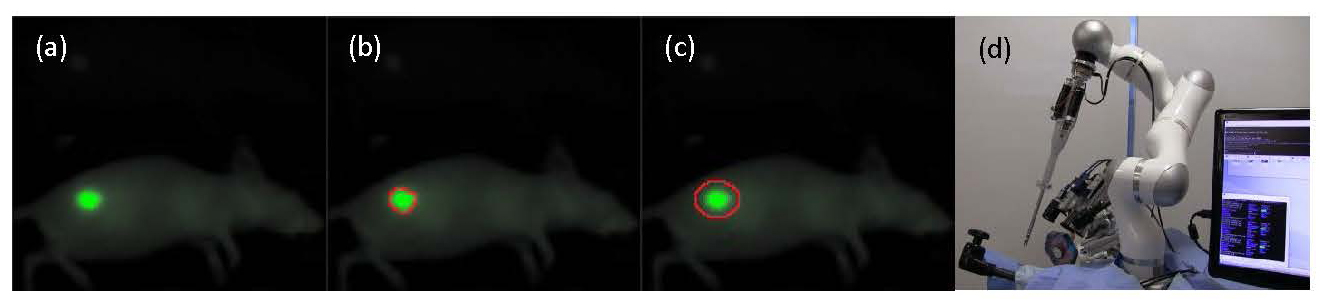
In this work, we will tag oral squamous cell carcinoma (OSCC) in the flank of an animal model with a novel near infrared tumor marking agent (FNIR) developed at the National Cancer Institute (Fig 1a). Tagged tumors will be imaged with a structured light camera, and segmented to identify tumor borders (Fig 1b). A path planning algorithm is used to generate an optimized surgical plan and margin for treatment (Fig 1c). Resection of the OSCC will be performed autonomously (STAR) (Fig 1d), manually (surgeon), and compared for margin consistency and percentage of cleared tumor bed.
Shedding New Light on the Inhibition of ATP-Binding Cassette Transporters
PRINCIPAL INVESTIGATORS:
Suresh Ambudkar, Laboratory of Cell Biology and Transport Biochemistry Section, Center for Cancer Research, National Cancer Institute
Huang Chiao Huang, Fischell Department of Bioengineering, University of Maryland
PH.D. STUDENT:
Barry Liang, Bioengineering, University of Maryland
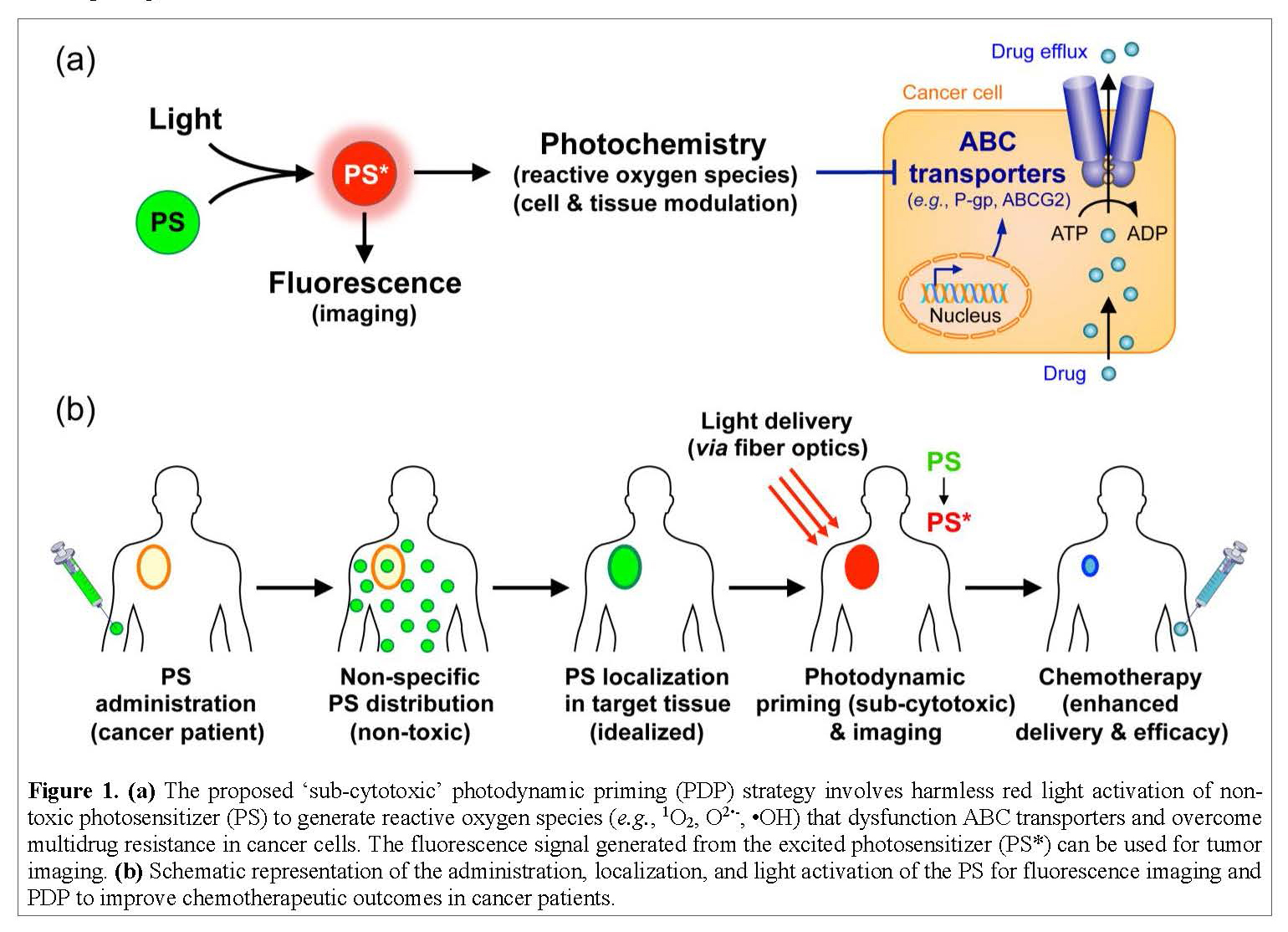
ATP-binding cassette (ABC) transporters are proteins found within our cells that take toxic chemicals from the inside of a cell and push them to the outside of a cell. Cancer adopts and amplifies this protective mechanism to pump the drugs out of the cell, thereby making them ineffective. Shutting down the ABC transporters only in cancer cells but not in normal tissues was impossible until now. An expert team of cancer biologist (Ambudkar), biomedical engineer (Huang), bioengineering Ph.D. student (Liang) and a CRTA student with a background in biochemistry (TBD) will study the mechanism by which light, chemistry, and nanotechnology can be exploited to shut down ABC transporters in cancer cells while sparing normal tissues.
Dual Proteomics–Transcriptomics of Developing Cranial Neural Crest Cells: New Frontiers for Cancer Research
PRINCIPAL INVESTIGATORS:
Ira Dear, Cancer & Developmental Biology Laboratory, National Cancer Institute
Peter Nemes, Department of Chemistry and Biochemistry, University of Maryland
PH.D. STUDENT:
Leena Pade, Chemistry, University of Maryland
Neural crest cells (NCCs) are stem-like cells that “borrow” migratory mechanisms from invading and metastasizing cancer cells to migrate significant distances as a loosely associated collective while differentiating into many tissues; therefore, determining how migratory NCCs interpret unique in vivo microenvironments in the developing vertebrate embryo raises a potential to identify new targets for inhibiting cancer metastasis in humans. However, our understanding of the underlying molecular mechanisms of abnormal cell migration and differentiation (cancer) has been hindered so far, because:
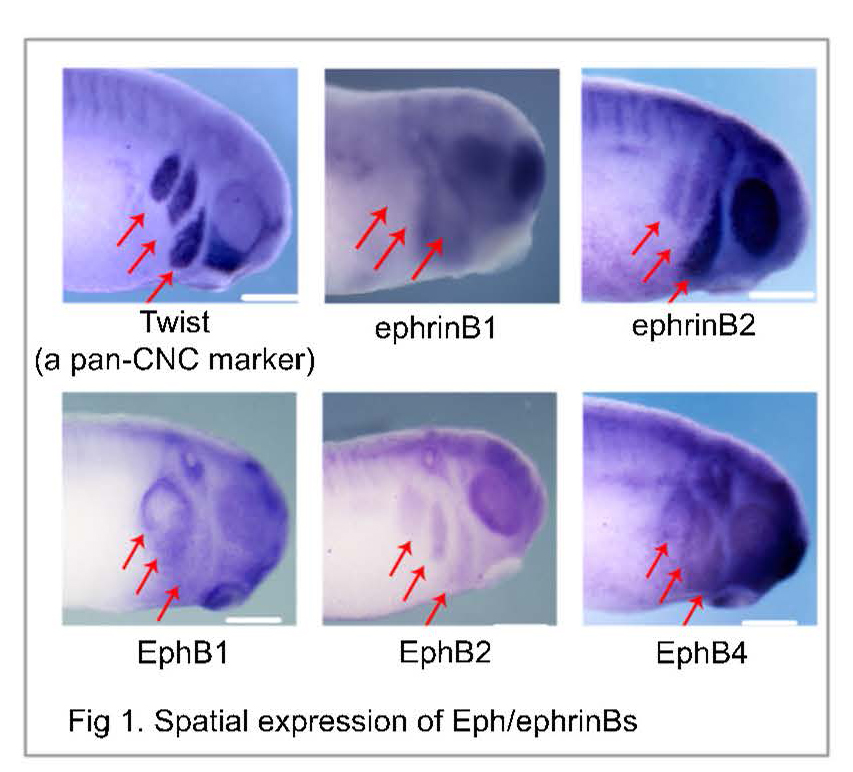
i) transcripts and proteins are poorly correlated in dynamic system, and so sequencing data by now routine Next-Gen Sequencing, qPCR, etc. alone provides incomplete information on functionally important proteins; and ii) the limited number of neural crest cells that can be dissected from embryos (e.g., Xenopus laevis, chick, zebrafish, mouse, etc.) contain too little amounts of proteins for characterization by commercial mass spectrometry, the modern technology of choice for the authoritative identification and quantification of hundreds-to-thousands of different proteins without using molecular probes (no antibodies). To fill this current knowledge gap in cancer research, we will perform the first dual transcriptomic-proteomic analysis of neural crest cells as they give rise to the four branchial (aka. pharyngeal) arches, each with a reproducibly different tissue fate, in Xenopus laevis (Daar Laboratory, NCI) using a custom-built microanalytical ultrahigh-resolution mass spectrometer capable of unprecedented sensitivity for proteins in single cells and small tissues in X. laevis and zebrafish embryos and mouse neurons (built and validated in the Nemes Laboratory, UMD). This work will effectively address cancer research at NCI by leveraging physical/personnel resources only available at UMD to allow the Daar and Nemes laboratories together (no prior collaboration) to generate new hypotheses using an important cancer model (X. laevis), thus forging a new UMD-NCI collaboration to seek external funding to advance cancer research forward (several FOAs via NIH, NSF, Foundations).
Understanding the Developmental Trajectory of CD8+ T Cells to Improve Immunotherapy
PRINCIPAL INVESTIGATORS:
Nick Restifo, Surgery Branch, Center for Cancer Research, National Cancer Institute
Eytan Ruppin, Cancer Data Science Laboratory, Center for Cancer Research, National Cancer Institute
Max Leiserson, Department of Computer Science, University of Maryland
PH.D. STUDENT:
Welles Robinson, Computer Science, University of Maryland
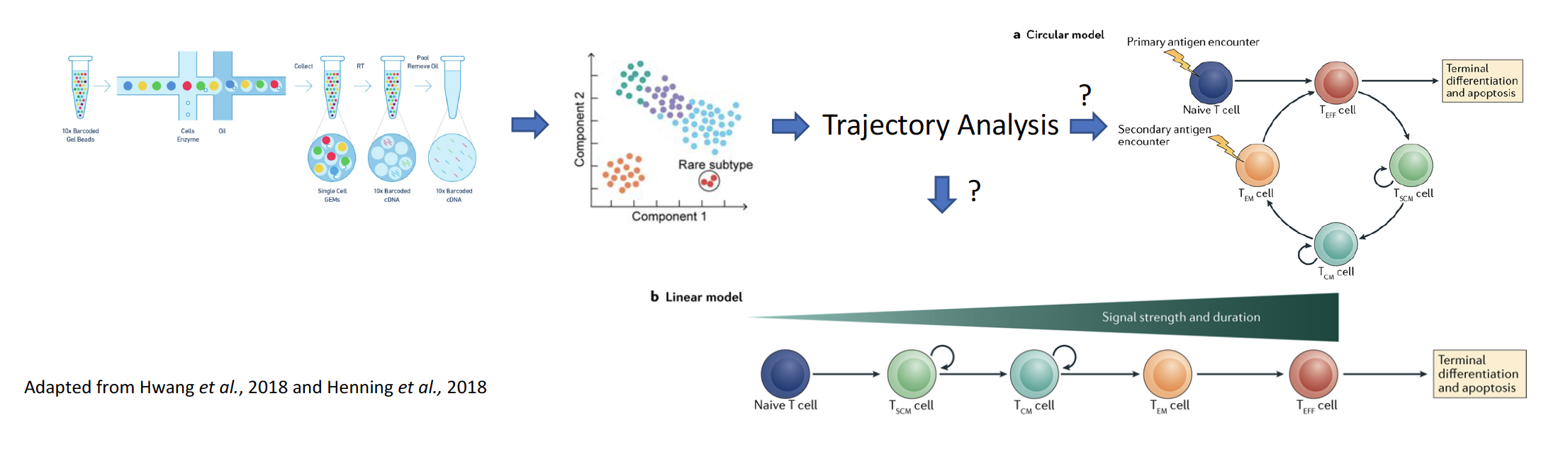
Stimulation of an immune response against cancer, including immune checkpoint blockade and adoptive transfer of tumor-infiltrating lymphocytes, has shown great promise in a limited number of cancer types. However, responses to adoptive cell transfer (ACT) are ‘anecdotal’ in solid epithelial cancers, which collectively account for almost 90% of the 600,000 cancer-related deaths in the US each year. An incomplete understanding of what makes CD8+T cells effective is one key barrier in the way of improving adoptive cell transfer (ACT), an important immunotherapy treatment pioneered by the NIH. To better understand the development of CD8+ T cells, we propose to reconstruct their developmental trajectory using coupled single cell RNA sequencing (scRNA-seq) and T cell receptor sequencing at an unpreceded, high resolution, which has the potential to resolve an important and fundamental question in T cell biology and impact patient treatment.
Understanding the Developmental Trajectory of CD8+ T Cells to Improve Immunotherapy
PRINCIPAL INVESTIGATORS:
Xin Wei Wang, Liver Cancer Section, Laboratory of Human Carcinogenesis, Center for Cancer Research, National Cancer Institute
Eric Slud, Department of Mathematics, University of Maryland
ASSOCIATE INVESTIGATORS:
Julian Candia, Liver Cancer Section, Laboratory of Human Carcinogenesis, Center for Cancer Research, National Cancer Institute
Doron Levy, Department of Mathematics, University of Maryland
PH.D. STUDENT:
Zeynep Kacar, Mathematical Statistics, University of Maryland
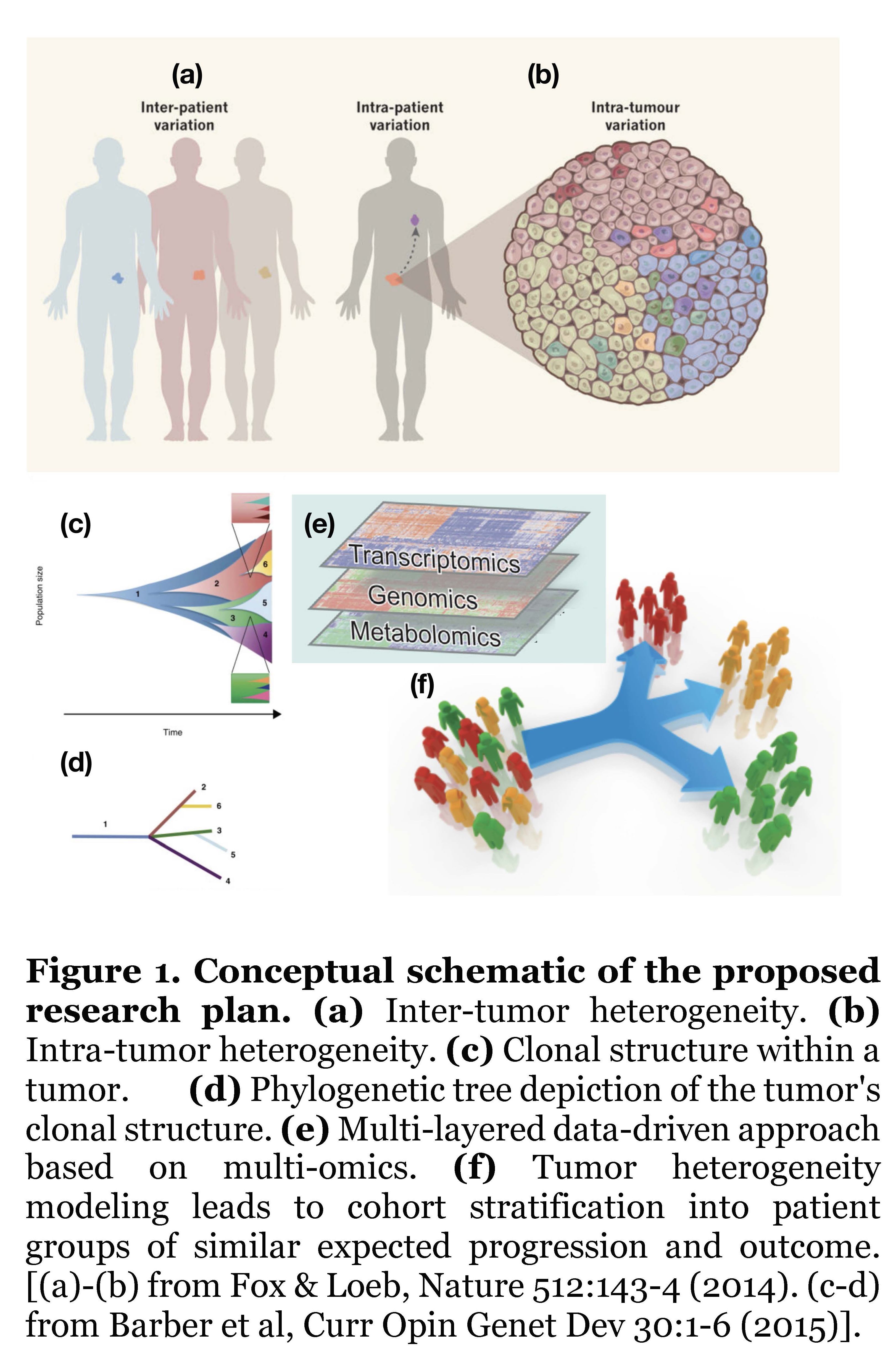
The goal of personalized cancer medicine is to recognize the important role of inter-patient variation (Fig.1(a)) to tailor treatment according to the molecular characteristics of each patient’s tumor; however, this rationale is hindered by the phenomenon of intra-patient variation (Fig.1(b)), whereby each patient’s tumor actually consists of multiple clones each carrying different sets of mutations. Based on mathematical and computational approaches applied to data from state-of-the-art sequencing technologies, it is possible to uncover the clonal structure and tumor history (Fig.1(c)), usually depicted in terms of phylogenetic trees, which are simplified structures representing the actual tumor clones (Fig.1(d)). By applying robust statistical models on different layers of molecular data (genomics, transcriptomics, metabolomics), this study will identify key driver mutations that promote tumor growth on those clonal structures (Fig.1(e)). The ultimate goal of our proposed research plan is to apply our mathematical models of tumor heterogeneity to stratify a cohort into patient subgroups of similar tumor progression and outcome (Fig.1(f)).
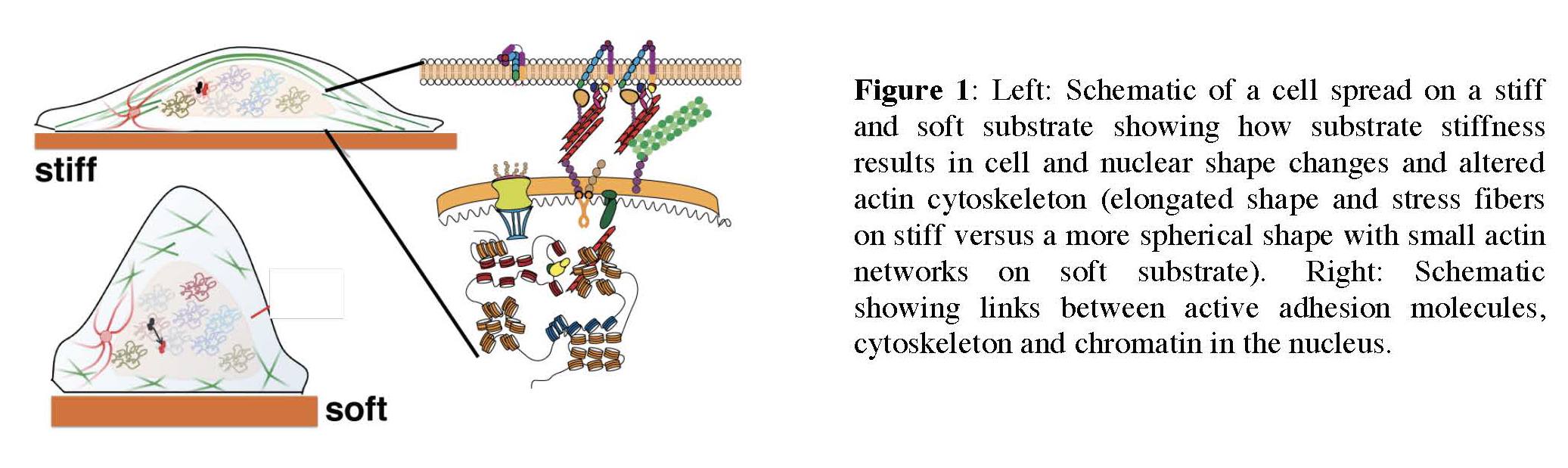
Mechanotransduction in the nucleus: how stiffness regulates transcription factor dynamics and gene expression
PRINCIPAL INVESTIGATORS:
Gordon Hager, Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, National Cancer Institute
Arpita Upadhyaya, Department of Physics, University of Maryland
PH.D. STUDENT:
Kaustubh Wagh, Physics, University of Maryland
Cellular responses to the physical properties of their surroundings such as stiffness, geometry and topography are critical to normal cell and tissue function as well as in cancer. However, how cells sense these mechanical signals and transduce them into long-term changes in gene expression that drive cellular decision-making is poorly understood. The goal of this research is to understand how changes in the mechanical environment of cells modulate transcriptional responses and gene expression. This research will combine the expertise from the Hager lab (NCI) and the Upadhyaya lab (UMD) to examine how the stiffness of the matrix on which cells are grown regulates nuclear organization and the dynamics of transcription factors in the nucleus, and has the potential to uncover fundamental mechanisms underlying mechanotransduction in cancer cells.