COMPUTATIONAL MODELING AND EXPERIMENTAL PERTURBATIONS OF THE CENTROMERIC CHROMATIN FIBER
PRINCIPAL INVESTIGATORS:
Yamini Dalal, Center for Cancer Research, National Cancer Institute
Garegin Papoian, Department of Chemistry and Biochemistry, University of Maryland
PH.D. STUDENT:
Haiqing Zhao, Biophysics, University of Maryland
Genomic DNA is intricately organized in cells of higher organisms, where histone proteins act as spools around which DNA winds, forming particles called nucleosomes, which further self-organize into fibrous structures called chromatin. Chromatin misregulation results in many human diseases, including cancers. In the proposed study, Dalal and Papoian laboratories will jointly investigate specific types of nucleosomes, where the H3 histones have been replaced by a variant called CENP-A, that serve as mechanical anchors for separating chromosomes during cell division. By mixing the structural domains of the H3 and CENP-A histones, the investigators will create and study both experimentally and computationally chimeric nucleosomal variants, that in turn will shed light on the way various CENP-A structural elements affect chromatin conformation and dynamics.
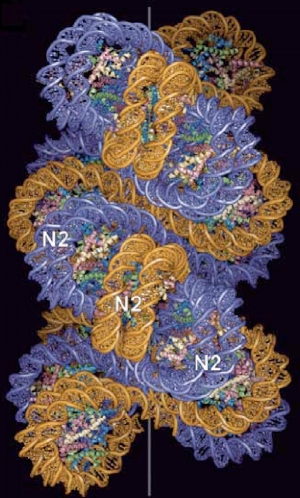
A proposed two step start structural model for compact chromatin fiber, reproduced from Richmond et al1. Histone tails are not shown.
CANCER CELL MRNA LOCALIZATION DURING METASTASIS
PRINCIPAL INVESTIGATORS:
Stavroula Mili, Center for Cancer Research, National Cancer Institute
Helim Aranda-Espinoza, Fischell Department of Bioengineering, University of Maryland
PH.D. STUDENT:
Susan Hamilla, Bioengineering, University of Maryland
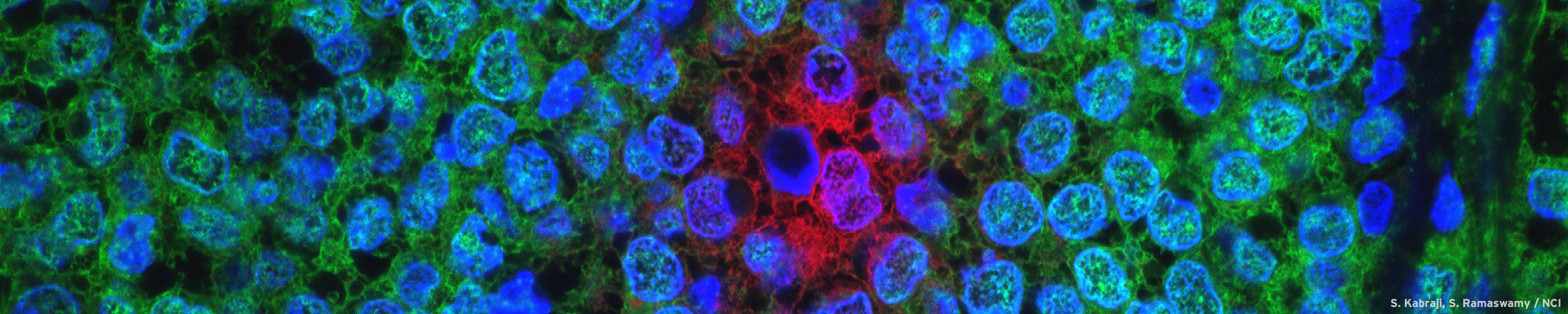
90% of cancer related deaths are caused by cancer metastasis. Localized mRNAs play an important role in cellular migration and localize to cellular protrusions and become disrupted in cancer progression. Additionally, cancer cell behavior is affected by the mechanical properties of the substrate and cancer cells modulate their gene expression in response to these extracellular cues. In this proposal, we would like to investigate mRNA localization in cancer cell protrusions during transmigration and on substrates of varying stiffness in order to gain mechanistic insights into cancer metastasis and its progression.
MDA-MB-231 breast cancer cells (red) transmigrating through a human umbilical vein endothelial cell monolayer (green) infected with a VE-Cadherin-GFP adenovirus. Scale bar is 10um.
ACCELERATING REAL-TIME CLINICAL DNA SEQUENCING DATA ANALYSIS WITH MASSIVELY PARALLEL COMPUTATION
PRINCIPAL INVESTIGATORS:
Javed Khan, National Cancer Institute
Uzi Vishkin, Department of Electrical and Computer Engineering, University of Maryland
Hector Corrada Bravo, Department of Computer Science, University of Maryland
PH.D. STUDENT:
Sean O'Brien, Electrical and Computer Engineering, University of Maryland
Genomic instability is a hallmark of most cancers and a major barrier for their effective treatment, making the complete characterization of unstable cancer genomes a prerequisite for the development of individualized therapies that effectively target currently incurable cancers. Our ability to use the vast amounts of data produced by next-generation sequencing (NGS) instruments hinges on having computational methods that can scale the analysis of raw sequence data so that results are generated within sufficient time to turn genomic insight into actionable courses of treatment. Our project seeks to significantly improve the efficiency and increase the speed of computational analysis of NGS data analysis to enable reporting of genomic alterations in a given data set. We will demonstrate the power of using the explicit multi-threaded (XMT) desktop supercomputer system architecture developed by co-PI Vishkin to accelerate sequence data analysis in a realtime clinical context by providing parallel implementations of local sequence assembly to discover chromosomal rearrangements and for variant detection.
ACCELERATED ELECTRON PARAMAGNETIC RESONANCE IMAGING FOR MEASURING CYCLING TUMOR HYPOXIA
PRINCIPAL INVESTIGATORS:
Murali Krishna Cherukuri, Center for Cancer Research, National Cancer Institute
Yu Chen, Fischell Department of Bioengineering, University of Maryland
Rao Gullapalli, University of Maryland School of Medicine
PH.D. STUDENT:
Chia-Chu Chou, Mathematics, University of Maryland
Compared to other competing imaging modalities, Electron paramagnetic resonance imaging (EPRI) is highly specific to tissue oxygenation and holds the potential of a low-cost and highly-specific modality to quantitatively and dynamically image tumor hypoxia in vivo and thus shows great potential for assessment of tumor physiology longitudinally to permit early evaluation of drug response. The focus of this proposal is to develop novel technologies that will accelerate EPRI acquisitions to enable these measurements to be made with greater spatial and temporal resolution. Development of novel image acquisition with high temporal resolution while maintaining superior spatial resolution will allow the observation of cycling hypoxia and lead to accurate measurement of intratumor tissue dynamics. Developments will allow tumor pO2 to be quantified spatially and temporally with previously unattainable spatio-temporal resolution and will provide new information on features such as erythocyte flux, tumor microvessel density, and the tumor microenvironment in general.
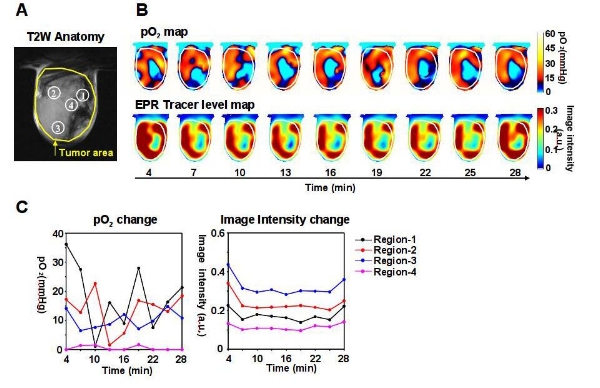
Example of a dynamic EPR imaging of a single 2D slice demonstrating tissue oxygenation (Fig 1) Figure 1(A) T2-weighted anatomical MR image of a tumor in the skeletal muscle of a mouse. (B) Dynamic images of calculated pO2 and EPRI signal intensity after tracer injection. (C) Temporal dynamics of pO2 and EPRI the regions shown in A.